Open OnDemand
From the comfort of your browser
Available on the BOSE supercomputing cluster to all users, Open OnDemand is an NSF-supported web-based platform that allows anyone to access their cluster resources right from any computer with a web browser. By not requiring the use of commands to interact with the cluster, this tool lowers the bar of entry for all students, faculty, and staff.
Note: If you are off campus, you will need to use the UWEC VPN before using Open OnDemand.
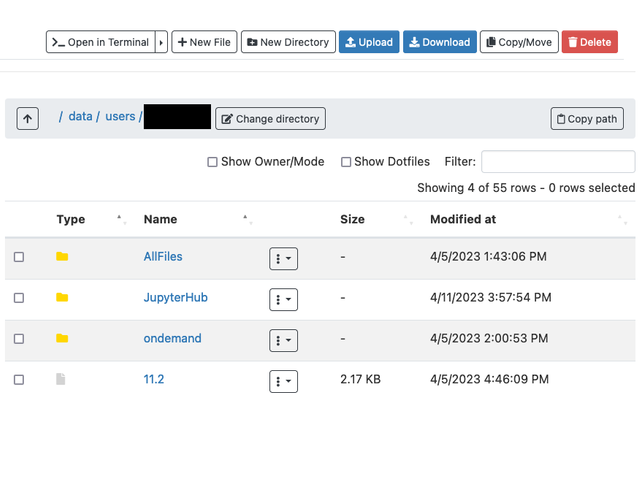
Accessing Files: Home Directory
Supported is the ability to fully manage, view, and edit all of your personal and group files right from OnDemand's built-in file browser.
Use "Open In Terminal" to easily jump into command mode for any additional work such as submitting a job.
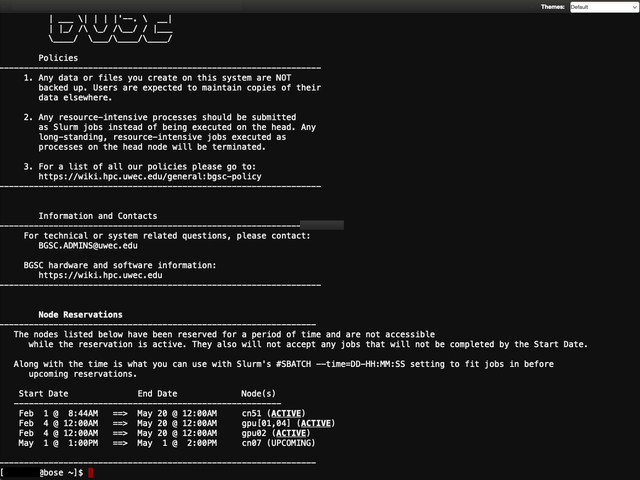
Cluster Shell Access
Available right in your browser window is the ability to log into BOSE as if you were using SSH through PuTTY, Command Prompt, or Terminal. From here, you are able to run any command, such as submitting jobs using sbatch.
Please note that using any GUI-based programs via X11 is not supported.
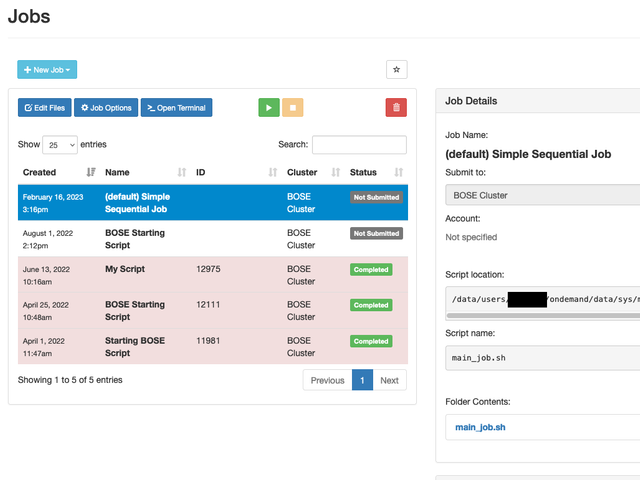
Job Composer
Instead of using the command-line to build and submit your Slurm scripts, you can use the Job Composer tool instead. This tool allows you to create new jobs based off existing ones, take advantage of script templates, modify submission files, and view your job's finished work.
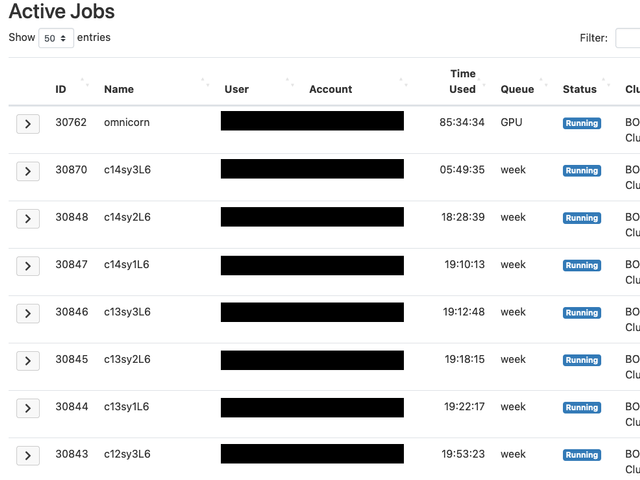
Active Jobs
Through the Active Jobs tool, you are able to view every job that is running on the cluster, as well as just your own.
This will let you see each resource that was requested, where it is running, and real time metrics of CPU and memory usage.
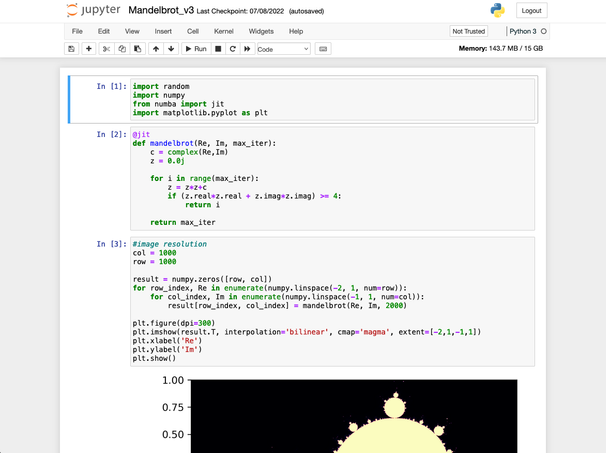
Jupyter Notebook
One of the most popular features of our BOSE cluster is our native integration with Jupyter. Used by both our researchers and in the classrooms, this tool allows everyone to spin up their own dedicated Jupyter server and interactively work with notebook files. This is a great way to work with Python code and view tables, graphs, and images in real time.
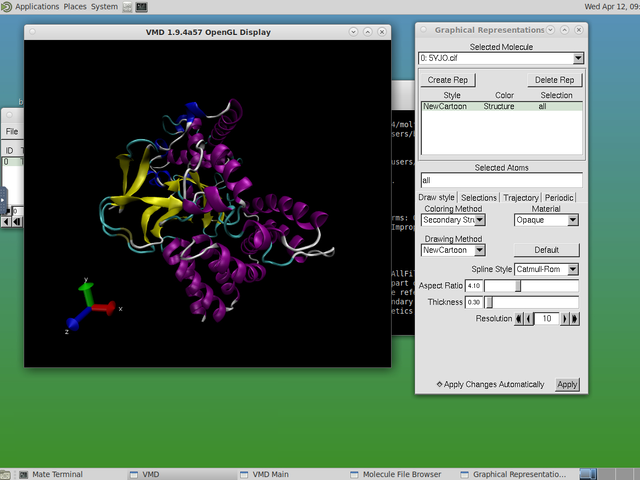
Desktop Mode
Desktop Mode allows you to access a compute node and work with it interactively as if it were a computer right in front of you. By using this tool, you can run desktop programs such as VMD (featured in screenshot) that require the use of both a mouse and keyboard to actively rotate and zoom 3D molecular structures.
Ready To Get Started?
Open OnDemand is a great resource to integrate into your class or research process. It lowers the bar of entry for utilizing our computing resources. Let us know you are interested in using OnDemand, and we will work alongside you to make sure everyone is up to speed. If interested, we are also available to provide training and presentations to your class or group.